New Solution: Cell cycle biosensors
Dave Matus presented the first technology session of the Embryo Journal Club on cell cycle biosensors on June 15th.
We discussed the usefulness of live cell cycle biosensors for understanding cell lineages and morphogenesis. First, Dave showed us a kind of cell cycle biosensor, also known as FUCCI, which was designed by a group in Japan (Sakue-Sawano et al, 2008). FUCCI, and several subsequent cell cycle biosensors like DHB (which shows CDK2 activity), enable us to visualize the dynamics of cell cycle using live samples, rather than fixed tissues.
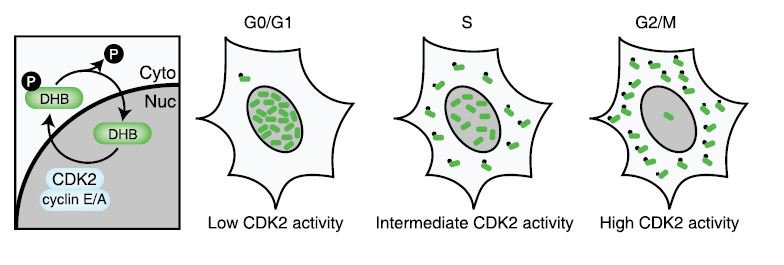
We talked about how the live cell cycle biosensors allow us to predict mitosis, and show the asymmetries in cell division: two daughter cells may look the same with a histone-gfp signal that simply marks the nuclei, but if the cells also have a cell cycle biosensor, their asynchronous cell cycles will become clear via the visual reporter.
Biosensors also help determining arrested/quiescent cells among a population of highly mitotic cells. In case of the specific cell type Dave studies (the anchor cell in C. elegans) the arrest was a key step in invasive behavior of the anchor cell: if not arrested, the anchor cell could not invade.
Readings
Spencer et al (2013) The Proliferation-Quiescence Decision Is Controlled by a Bifurcation in CDK2 Activity at Mitotic Exit
Matus et al (2015) Invasive Cell Fate Requires G1 Cell-Cycle Arrest and Histone Deacetylase-Mediated Changes in Gene Expression


Comments
Post a Comment